Evaluating the Medical Knowledge of Open LLMs - Part 1
By Tanishq Mathew Abraham, Ph.D. and Griffin Adams
TL;DR: SOTA open generalist LLMs like Qwen-72b perform better than Med-PaLM on the MultiMedQA suite of tasks without any finetuning or even prompt engineering, closing the gap between proprietary and open LLMs for medical tasks.
If this sort of research interests you and you have experiences fine-tuning/training LLMs, we're hiring! Apply here
Introduction
Large language models (LLMs) have been successfully applied to a diverse set of tasks, ranging from coding assistants to robotics. One area of keen interest is healthcare, where LLMs have the potential to assist care providers with administrative workflows and to provide clinical decision support. As such, there has been significant research in the past two years related to both understanding the medical capabilities of LLMs, as well as developing LLMs trained on medical text. Generalist LLMs like GPT-4 and PaLM exhibit medical knowledge that can be elicited via prompt engineering or minimal finetuning. While this approach has often resulted in state-of-the-art (SOTA) performance, training of medical domain-specific LLMs has also shown significant promise and it is an open question which approach is superior.
At MedARC, we are interested in studying this question in further detail and developing the best open LLMs for medical tasks. We believe the release of domain-specialized open-source LLMs is crucial for practical medical applications of the latest LLM advances. While there has been some work done in the field, like BioMedLM, Clinical Camel, Meditron, etc., there is still plenty of work to be done in the area.
In order to develop the best open medical LLMs, we need to be able to evaluate our progress. Currently, the evaluation of LLMs on medical tasks is most commonly done on MultiMedQA, a suite of medical question-answering tasks introduced by Google in the Med-PaLM paper. MultiMedQA includes tasks like PubMedQA, MedQA, MMLU clinical knowledge, etc., which evaluate medical knowledge and reasoning. As of January 2024, prompt-engineered GPT-4 is SOTA on all tasks in MultiMedQA.
While closed-source generalist LLMs like PaLM and GPT-4 have been thoroughly evaluated on medical tasks in MultiMedQA and on other benchmarks, there has been minimal evaluation of generalist open LLMs on medical tasks. Benchmarking open source LLMs is the first step toward selecting which models to use and possibly fine-tune for medical applications. Therefore, we evaluate a variety of SOTA generalist open LLMs (Llama-2, Mistral, Yi, etc.) on MultiMedQA to see how well they stack up against proprietary LLMs.
Methodology
We have implemented MultiMedQA within EleutherAI's lm-eval-harness and have added it to the main library. lm-eval-harness is a popular library for evaluating language models, and we hope the addition of the MultiMedQA suite to lm-eval-harness will enable the community to better study and understand the medical capabilities of LLMs.
MultiMedQA is comprised of 9 multiple-choice question answering tasks:
PubMedQA - 1,000 expert-labeled Q&A pairs where a question and corresponding PubMed abstract as context is given, and a yes/maybe/no answer must be produced. Unlike the rest of the tasks in this suite, PubMedQA is a closed-domain Q&A task.
MedQA - US Medical License Exam (USMLE) questions with 4 or 5 possible answers. Typically, only the 4-option questions are used.
MedMCQA - 4-option multiple choice questions from Indian medical entrance examinations, >191k total questions.
MMLU - 4-option multiple choice exam questions from a variety of domains. The following six domains are utilized here:
Anatomy
Clinical Knowledge
College Medicine
Medical Genetics
Professional Medicine
College Biology
Note that MultiMedQA also includes some short-form and long-form Q&A tasks (LiveQA, MedicationQA, HealthSearchQA). Evaluation of these tasks is usually done by experts and is not typically performed automatically and therefore is ignored here.
We have evaluated a total of 22 models on MultiMedQA. This includes models like Llama-2, Yi, Mistral, StableLM, and Qwen models. This also includes one domain-specific fine-tuned model, Meditron-70b, which is one of the best open-source medical LLMs. We perform both zero-shot and five-shot evaluation for each model. The evaluation approach as done in lm-evaluation-harness is that for each task, the conditional log likelihood of each answer letter (A, B, C, D) given the document is calculated, and the answer choice with the highest log-likelihood is selected. In the case of PubMedQA, the length-normalized log-likelihood for the "yes", "no", and "maybe" answer options are compared.
Results
The below table reports the best score for each task, what model it was, and whether it was zero-shot or five-shot:
Table 1: A comparison of the best open LLMs to current SOTA on MultiMedQA
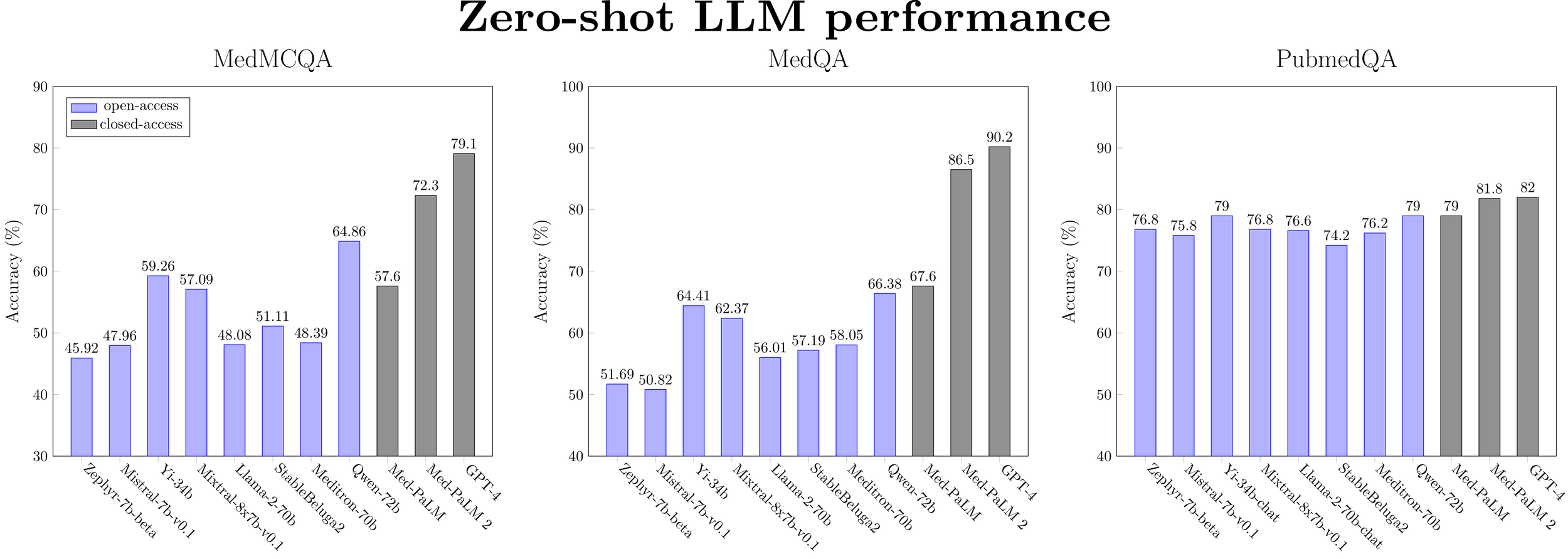
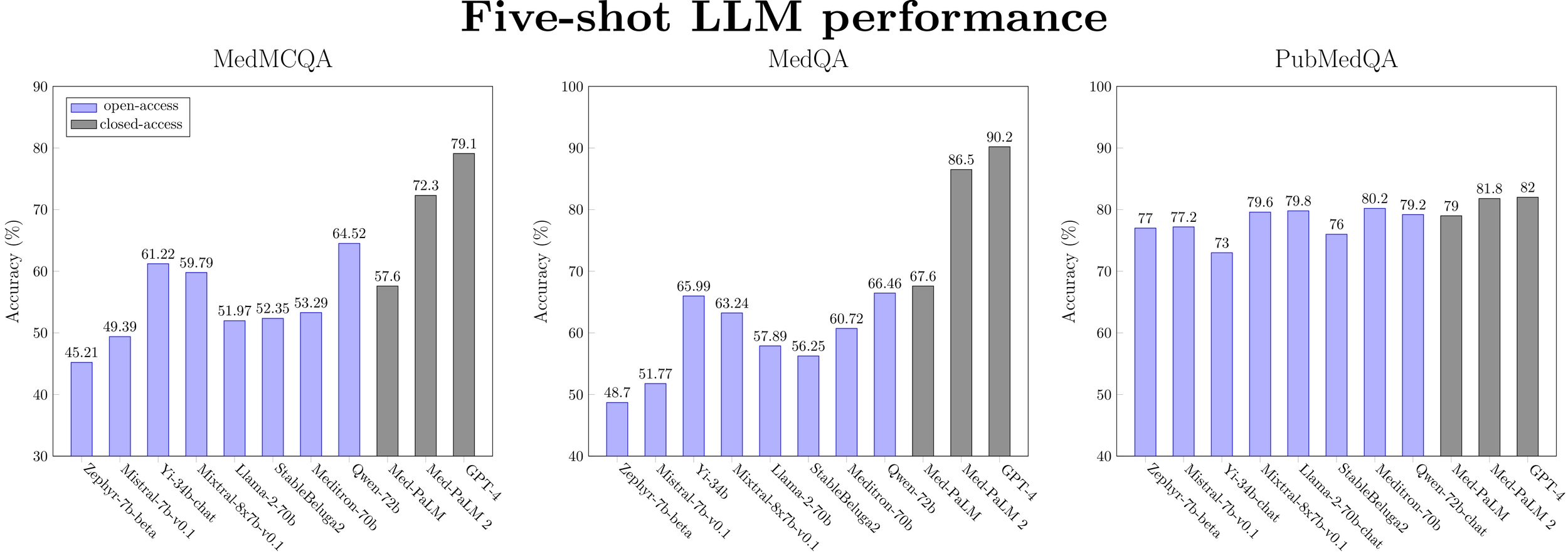
Overall, there is a clear winner: Qwen-72b is the best open medical LLM on medical reasoning benchmarks in the zero-shot setting. It is within 9.7% of Med-PaLM 2’s performance and 13.8% of GPT-4’s performance. On some tasks, open models are closing in on the SOTA (PubMedQA, MMLU college biology). However, current open base models still remain inferior to GPT-4 and Med-PaLM 2 but are better than Med-PaLM on all tasks except MedQA. Note that Med-PaLM, Med-PaLM 2, and GPT-4 use sophisticated prompting techniques, while we only test simple zero- or five-shot prompting.
Given this context, the performance of these open models is arguably quite impressive. As Med-PaLM was presented at the end of 2022, a reasonable conclusion is that open LLMs are at most one year behind proprietary LLMs. Of course, sophisticated prompting techniques can be applied to the open LLMs just as they were applied to proprietary LLMs and the gap between open LLMs and the SOTA with proprietary models may actually be smaller.
Let's take a deeper dive into the results and any trends we observe. The full table of results is available here.
Figure 1: Zero-shot performance for a selected subset of LLMs (ranked by model size) on medical QA datasets.
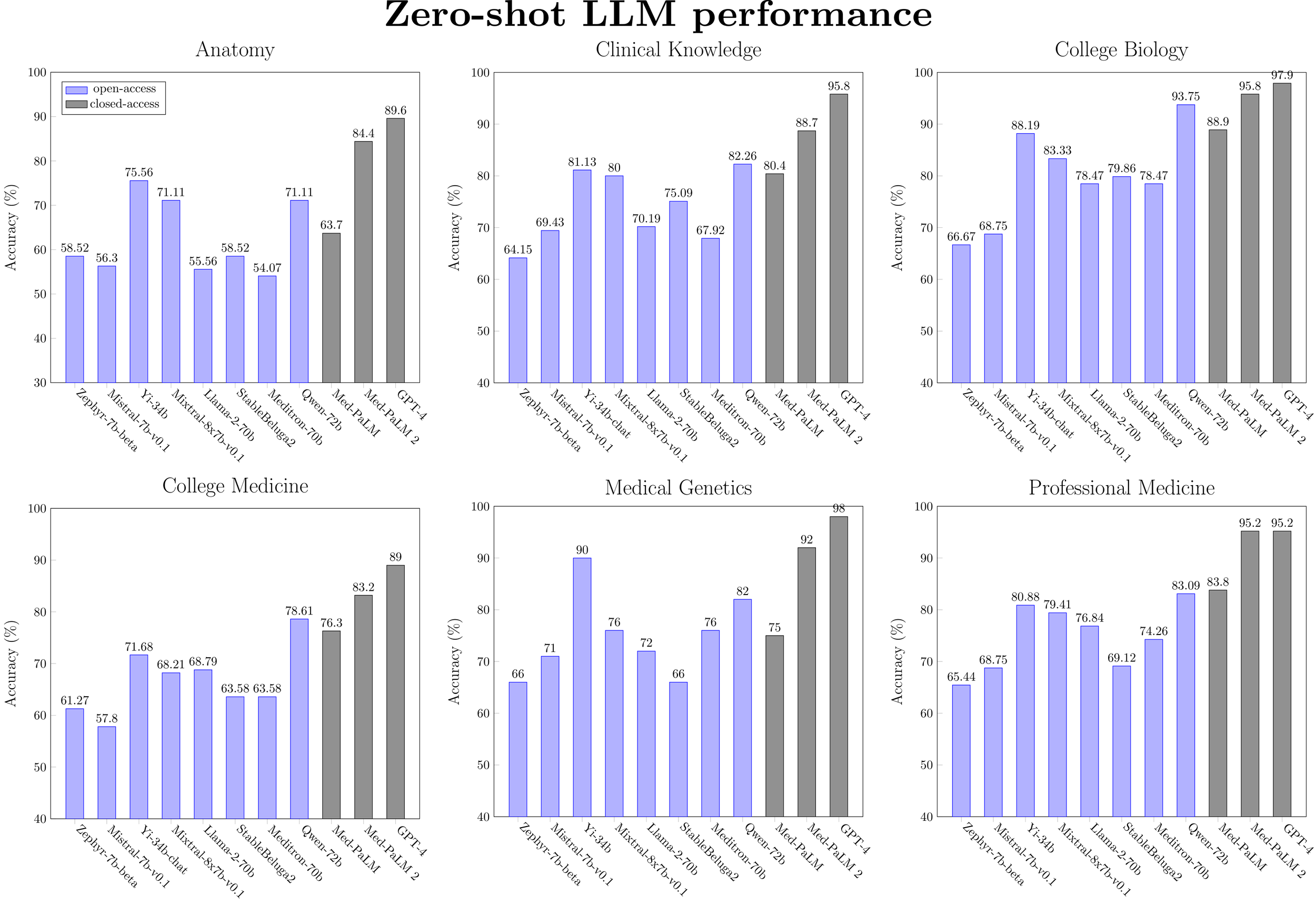
Figure 2: Zero-shot performance for a selected subset of LLMs (ranked by model size) on medical MMLU tasks.
Figure 3: Five-shot performance for a selected subset of LLMs (ranked by model size) on medical QA datasets.
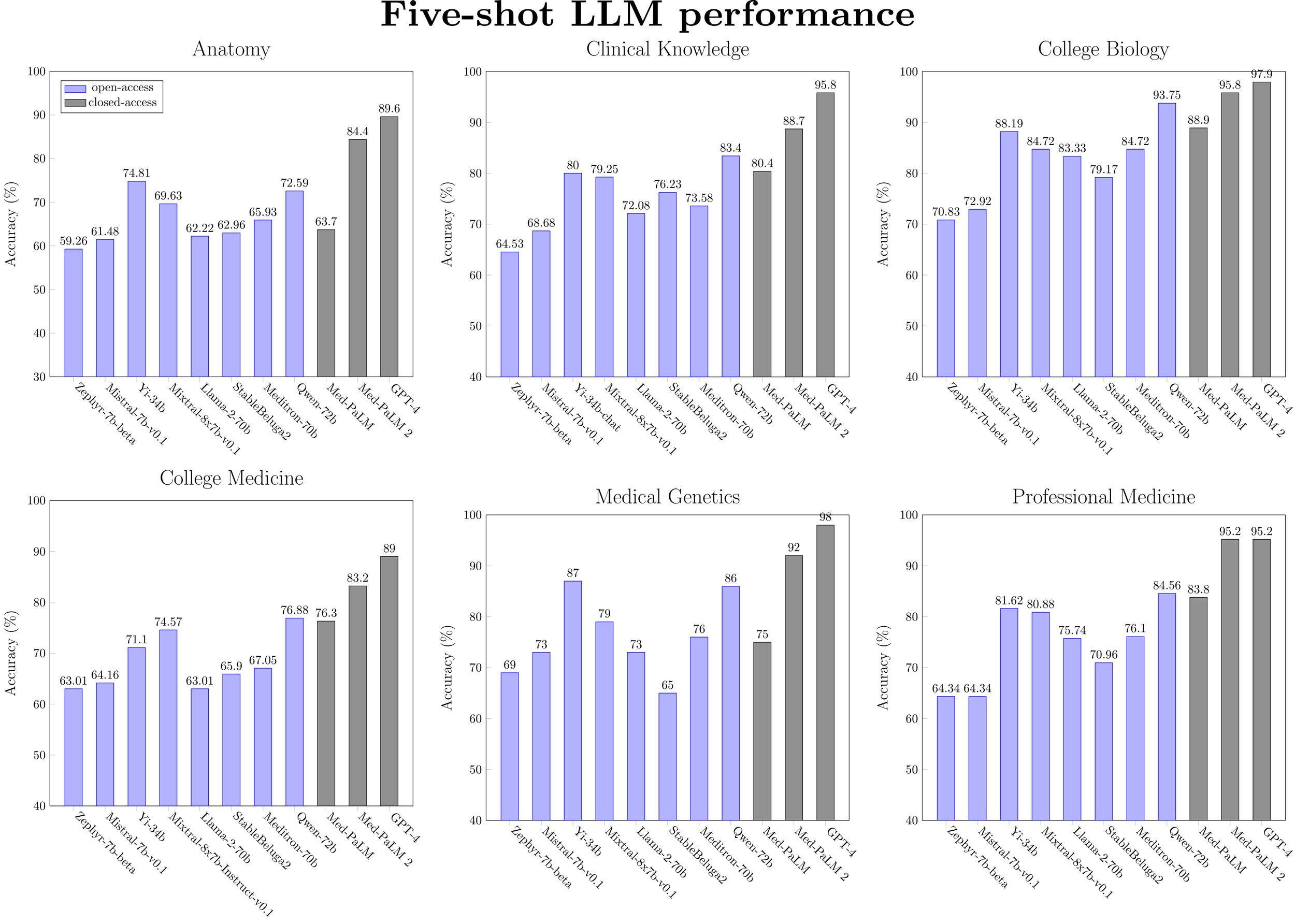
Figure 4: Five-shot performance for a selected subset of LLMs (ranked by model size) on medical MMLU tasks.
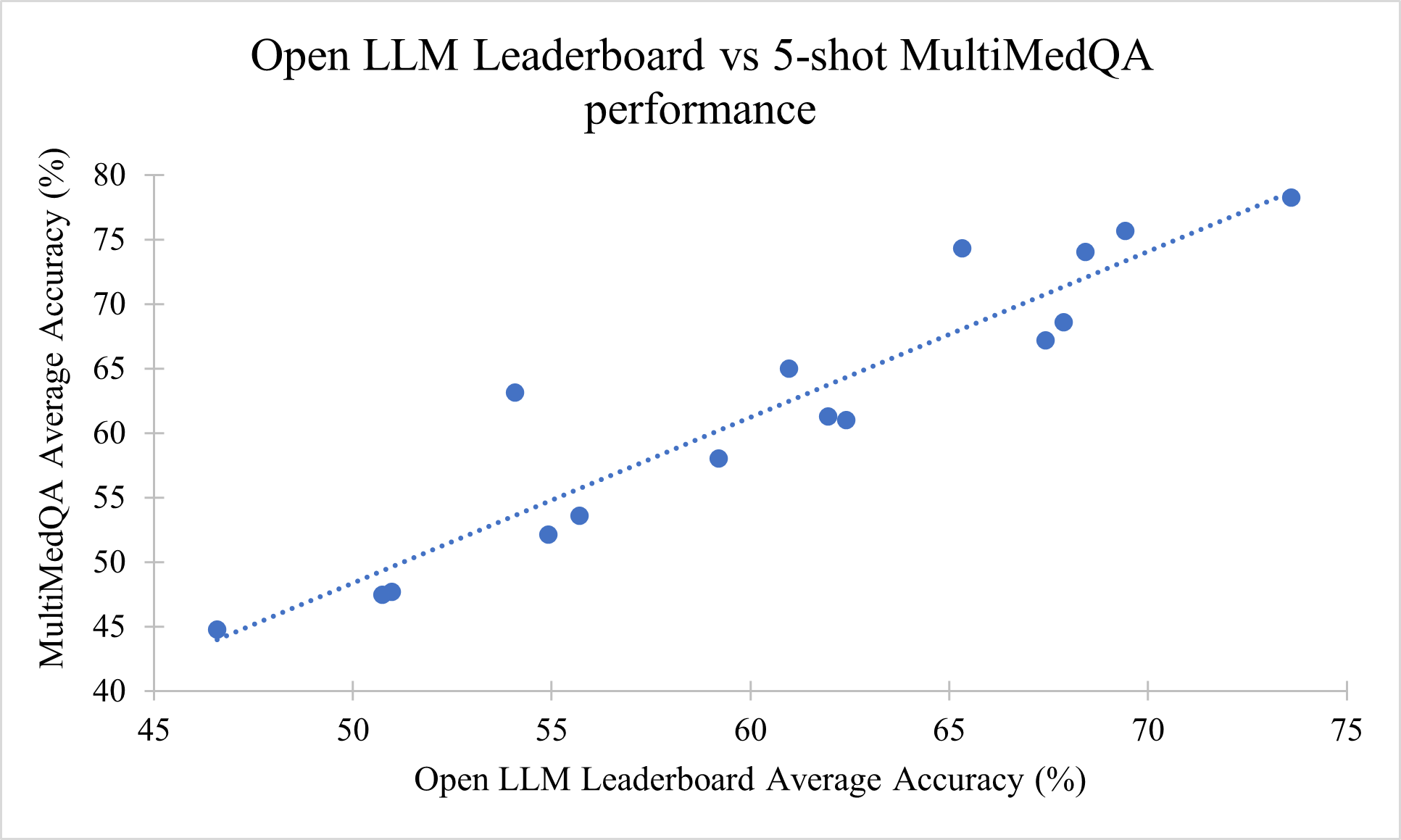
Figure 5: Correlation between Open LLM Leaderboard performance and five-shot MultiMedQA performance (R2 =0.8802).
Medical capability (as measured by MultiMedQA) is correlated with general model performance (as measured by Open LLM Leaderboard). The best open base models as of Jan 2024 (Yi-34b and Qwen-72b) that are outperforming Med-PaLM, despite no specific training for medical tasks.
Llama-2 models are not close to state-of-the-art for medical knowledge. This is true both generally and even within their respective model size classes. Apart from on the PubMedQA task, Llama-2-7b is beat significantly by Mistral-7b, Yi-6b and Qwen-7b, while Llama-2-70b is beat by Yi-34b, Mixtral-7b (46.7B params), StableBeluga2, and Qwen-72b. Most recent work into developing open medical-specific LLMs has focused on finetuning or continued pretraining of Llama-2 models. For example, Meditron-70b was obtained through continual pretraining of Llama-2-70b on PubMed and medical guidelines. However, on the MultiMedQA benchmark, we observe that Meditron-70b is inferior to the Yi-34b and Qwen-72b generalist models. We hypothesize that finetuning these other classes of generalist LLMs would result in better performance.
Chat models do not provide a consistent benefit in performance over base models. Apart from a minor performance gain observed with Llama-2-7b and Yi-6b models (i.e. the smaller-scale models), the zero-shot performance of instruction-tuned models is similar or worse than the base models. For five-shot performance, no benefit is observed at any model scale. This surprising finding warrants further investigation, which is outside the scope of this blog post.
Dataset contamination
Given the impressive performance of generalist models like Yi-34b and Qwen-72b, a natural question is whether there has been any dataset contamination, i.e., evidence of “training on the test.”
Due to the overall lack of transparency surrounding the pretraining datasets of most open LLMs, we perform contamination analyses using five recently proposed methods for Membership Inference Attacks (MIAs). Each attack seeks to answer the question:
Given black-box access to an LLM, was the model trained on any of the test sets in MultiMedQA?
MELD: This was proposed in Capabilities of GPT-4 on Medical Challenge Problems. It involves bifurcating each test question in half and prompting each model to finish the question. Similarity between the model-generated completion and the actual completion is measured with the inverse of the length-normalized Levenshtein distance. If the similarity is >= 0.95, this test question was likely in the training set.
Min-K% Prob: This was proposed in Detecting Pretraining Data from Large Language Models. For each test set example, it involves computing the token-level log-likelihoods. The likelihoods are then sorted, and then the lowest K-% are averaged to produce a score. A Min-K% Prob > ε suggests contamination. They tune ε = -7.3523 on the validation set of a newly proposed MIA benchmark (WikiMIA).
Quiz Accuracy: This was proposed in Data Contamination Quiz: A Tool to Detect and Estimate Contamination in Large Language Models A Tool to Detect and Estimate Contamination in Large Language Models. The authors design a multiple-choice quiz which is intended to get the model to “out” itself. For each test case, they generate three paraphrases with GPT-4. They then randomly insert the original test case into the list of paraphrased examples. The LLM is then asked to pick out the real test case from the paraphrases. The level of contamination is viewed as positively correlated to the chance-adjusted cohen’s kappa for the quiz accuracy.
Neighborhood Loss Delta: This was proposed in Membership Inference Attacks against Language Models via Neighbourhood Comparison. We take the same paraphrase data constructed for the above quiz and compute the average log likelihoods. The lower the difference between the log likelihood on the original and the average log likelihood of the paraphrases, the more likely the model is contaminated. Without a given threshold for classification, we simply report outlier deltas (< 0.5) below.
Guided Instruction: This was proposed in Time Travel in LLMs: Tracing Contamination in Large Language Models. Using two different instructions, we instruct each LLM to generate a question for each test set input, given the inputs and target answers. The first set of instructions (“Guided”) instructs the model to generate the question as it appears in the specific test set. The second instruction (“General”) simply asks the model to generate any question. We compute the ROUGE-L score between the model generated questions and the original questions for both sets of instructions. If the ROUGE-L for “Guided” is statistically significantly higher (p < 0.05), the model may be contaminated.
We compute these five scores across each test set in MultiMedQA across 4 of the top-performing models (Yi-34b, Mixtral-7b, Llama-2-70b, Qwen-72b) and report only on any outliers or potential evidence of contamination.
MELD: No evidence
Min-K% Prob: Nearly all the models fail according to the Min-K% Probability test using the threshold tuned by the authors on a newly created detection benchmark called WikiMIA. To investigate further, we would likely need to tune our own thresholds using MultiMedQA data.
Quiz Accuracy:
PubMedQA: Yi-34b has a relatively high Cohen’s Kappa score of 0.325
Neighborhood Loss Delta
PubMedQA: Yi-34b and Qwen-72b have relatively low Neighborhood Loss Deltas of -0.592 and -0.526, respectively.
MMLU - Professional Medicine: Yi-34b has a low Neighborhood Loss Delta of -0.715
Guided Instruction: No evidence. Most models struggled to recreate the test set questions with ROUGE-L scores mostly <= 20 across datasets.
This analysis merely suggests that Yi-34b might be exposed to some of the datasets in MultiMedQA, as it was flagged by 2 metrics (Quiz Accuracy and Neighborhood Loss Delta). Yet, for PubMedQA at least, Yi-34b was not flagged by MELD and Guided Instruction, so the results are not conclusive.
Additionally, we do not go so far as to say that Yi-34b’s results are not valid. To this effect, we want to emphasize two important caveats.
False positives may exist. These methods for blackbox MIA detection are not bulletproof, especially those which rely on synthetic (GPT-4) paraphrases. Models which are better able to detect synthetic data from real data might be unfairly penalized by these methods.
Models may memorize contaminated data but not exploit it (use it to improve performance). In short, evidence of contamination does not necessarily mean that a model is worse than it appears.
Conclusion
In conclusion, we have preliminarily shown that current generalist open LLMs appear to have medical capabilities that are better than Med-PaLM with no special prompt engineering or fine-tuning. We have identified Qwen-72b specifically as the current best-performing open model. These generalist models even beat some models trained specifically for improved medical knowledge (Meditron-70b). Note, however, that these domain-specific models were often finetuned off the Llama series of models or older models or even trained from scratch. Therefore, this does not necessarily indicate that generalist models are all you need. Instead, this indicates an opportunity to instead finetune the best-performing models like Qwen-72b in hopes of obtaining even better performance in the medical domain.
By integrating our MultiMedQA assessment into the lm-eval-harness, we aim to ensure the ongoing tracking of the medical capabilities of open LLMs as the field continues to grow.
In future blog posts, we'll study the application of prompt engineering techniques to improve model performance and also task-specific finetuning as another axis to evaluate a model's medical capabilities.
Acknowledgements
Thank you to Jean-Benoit Delbrouck and Katie Link for initial contributions for the lm-eval-harness implementation of MultiMedQA. Thank you to Paul Scotti, Nathan Cooper, Jeremy Howard, and Hailey Schoelkopf for feedback and discussions.
To cite this blog post, please use:
T.M. Abraham, G. Adams. (Jan 2024). Evaluating the Medical Knowledge of Open LLMs - Part 1. MedARC Blog. https://medarc.ai/blog/medarc-llms-eval-part-1
@article{abraham2024llmeval,
title="Evaluating the Medical Knowledge of Open LLMs - Part 1",
author="Abraham, Tanishq Mathew and Adams, Griffin",
journal="MedARC Blog",
year="2024",
month="January",
url="https://medarc.ai/blog/medarc-llms-eval-part-1"
}